msDiaLogue is a customized, modular, and flexible workflow developed jointly by UConn’s PMF and SCS for data-independent acquisition (DIA) mass spectrometry (MS)-based proteomics data.
Modular Structure
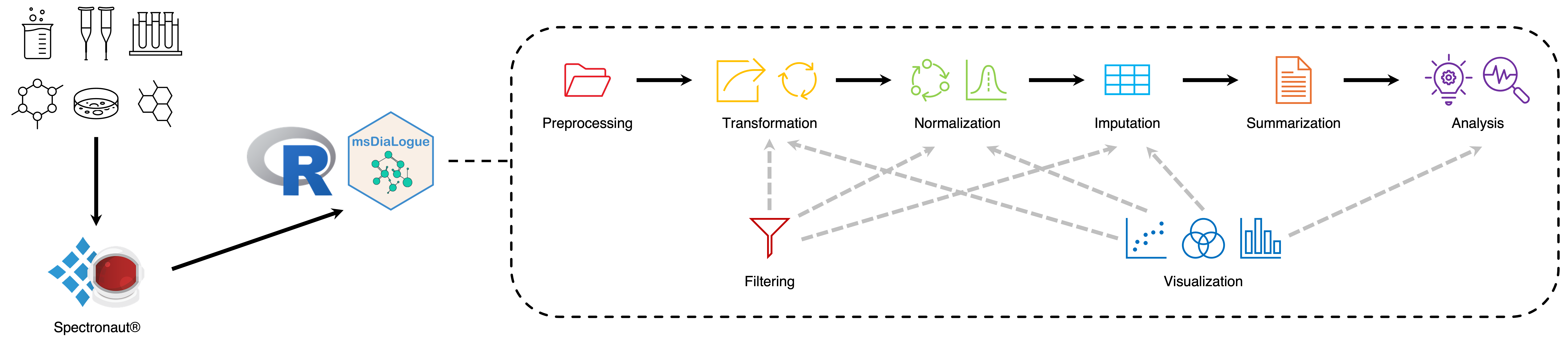
- Preprocessing: Getting data from Spectronaut.
- Transformation: Options for transforming abundance data.
- Normalization: Normalization procedures.
- Imputation: Missing data procedures.
- Summarization: Calculating and presenting numerical summaries in tabular form.
- Analysis: Statistical tools for DIA data analysis.
- Filtering: Providing options to filter out data based on preset levels.
- Visualization: Providing clean visuals to aid in data analysis decisions.
Installation
You can install msDiaLogue directly from GitHub.
- Development (latest) version:
# install.packages("devtools")
devtools::install_github("uconn-scs/msDiaLogue")- Specific released version (e.g. v0.0.6):
# install.packages("devtools")
devtools::install_github("uconn-scs/msDiaLogue@v0.0.6")