Preprocessing
Shiying Xiao, Charles Watt, Jennifer C. Liddle, Jeremy L. Balsbaugh, Timothy E. Moore
Department
of Statistics, UConn
Proteomics
and Metabolomics Facility, UConn
Statistical
Consulting Services, UConn
2025-11-25
Source:vignettes/preprocessing.Rmd
preprocessing.RmdPreliminary
## load R package
library(msDiaLogue)Example
## if the raw data is in a .csv file
fileName <- "../inst/extdata/Toy_Spectronaut_Data.csv"
dataSet <- preprocessing(fileName,
filterNaN = TRUE, filterUnique = 2,
replaceBlank = TRUE, saveRm = TRUE)preprocessing() does not perform a
transformation on your data. You still need to use the function
transform().
## if the raw data is in an .Rdata file
load("../inst/extdata/Toy_Spectronaut_Data.RData")
dataSet <- preprocessing(dataSet = Toy_Spectronaut_Data,
filterNaN = TRUE, filterUnique = 2,
replaceBlank = TRUE, saveRm = TRUE)
#> Warning: Removed 62 rows containing non-finite outside the scale range
#> (`stat_bin()`).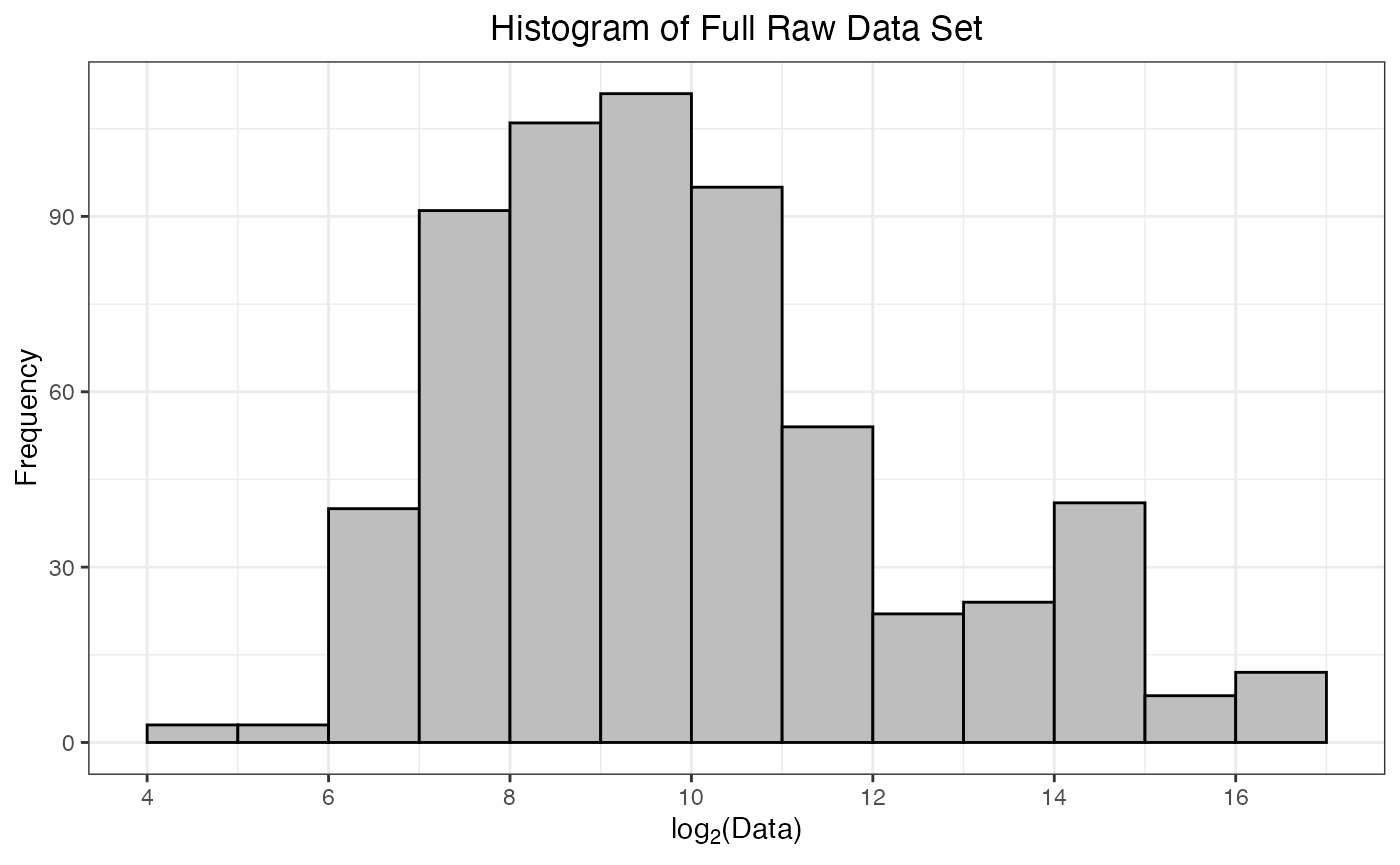
#> Summary of Full Data Signals (Raw):
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> 20.93 263.87 669.79 6897.92 1963.53 117803.49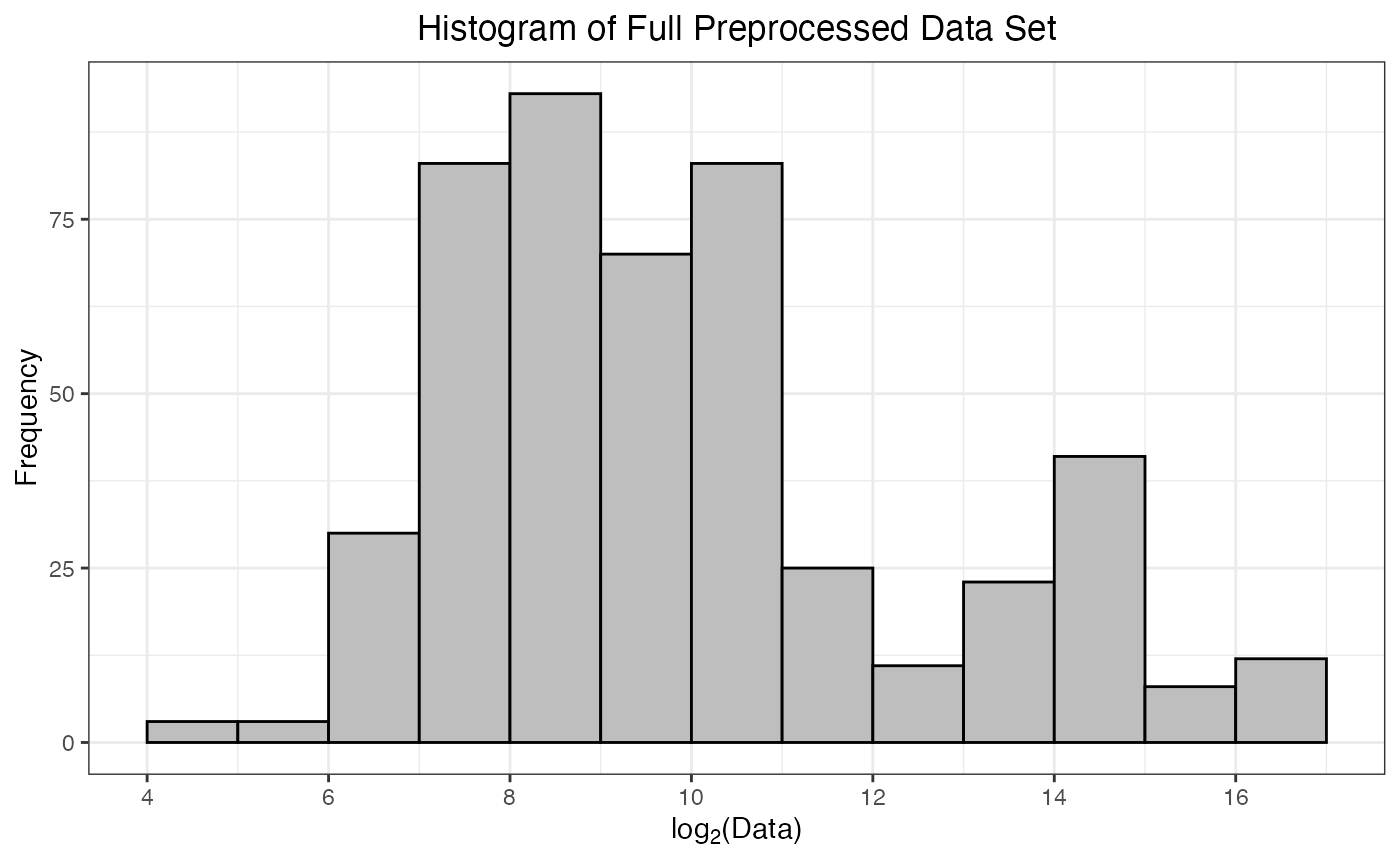
#> Levels of Condition: 100pmol, 200pmol, 50pmol
#> Levels of Replicate: 1, 2, 3, 4| R.Condition | R.Replicate | NUD4B_HUMAN (+1) | A0A7P0T808_HUMAN (+1) | A0A8I5KU53_HUMAN (+1) | ZN840_HUMAN | CC85C_HUMAN | TMC5B_HUMAN | C9JEV0_HUMAN (+1) | C9JNU9_HUMAN | ALBU_BOVIN | CYC_BOVIN | TRFE_BOVIN | KRT16_MOUSE | F8W0H2_HUMAN | H0Y7V7_HUMAN (+1) | H0YD14_HUMAN | H3BUF6_HUMAN | H7C1W4_HUMAN (+1) | H7C3M7_HUMAN | TCPR2_HUMAN | TLR3_HUMAN | LRIG2_HUMAN | RAB3D_HUMAN | ADH1_YEAST | LYSC_CHICK | BGAL_ECOLI | CYTA_HUMAN | KPCB_HUMAN | LIPL_HUMAN | PIP_HUMAN | CO6_HUMAN | BGAL_HUMAN | SYTC_HUMAN | CASPE_HUMAN | DCAF6_HUMAN | DALD3_HUMAN | HGNAT_HUMAN | RFFL_HUMAN | RN185_HUMAN | ZN462_HUMAN | ALKB7_HUMAN | POLK_HUMAN | ACAD8_HUMAN | A0A7I2PK40_HUMAN (+2) | NBDY_HUMAN | H0Y5R1_HUMAN (+1) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 100pmol | 1 | 1547.983 | 3168.32568 | 2819.7874 | 318.54376 | 495.5136 | 456.3309 | 213.21727 | 237.1306 | 111209.7 | 10737.953 | 15097.67 | 1799.391 | 630.1937 | 1311.8127 | 1279.6390 | 280.6318 | 299.51523 | 1154.5566 | 16461.2012 | 179.3190 | 516.1104 | 1234.587 | 27599.42 | 13798.590 | 23840.03 | 614.0895 | 990.5613 | 440.0417 | 132.31737 | 150.6033 | 3578.014 | 26872.50 | 109.55331 | 211.6450 | 1292.5234 | 1963.5321 | 189.79155 | 1106.1482 | 981.11432 | 180.6320 | 199.14555 | 209.7806 | NA | NA | NA |
| 100pmol | 2 | 1680.730 | 4576.37158 | 1061.9502 | 404.25836 | 556.8611 | 501.0473 | 184.89574 | 314.0320 | 111659.9 | 10655.384 | 15840.28 | NA | 575.0490 | 1114.2773 | 1294.9751 | 271.8160 | 248.04329 | 1032.0381 | 1460.7496 | 213.1137 | 492.3771 | 1186.433 | 27221.59 | 13880.411 | 23963.31 | 640.2153 | 1077.4829 | 364.5241 | 128.78983 | 128.2592 | 3412.794 | 26742.22 | 155.37483 | 348.6104 | 1066.3511 | 1509.1512 | 153.90802 | 1303.6520 | 388.65823 | 122.7458 | 751.19849 | 247.3832 | 1420.1351 | NA | NA |
| 100pmol | 3 | 1414.811 | 4675.13281 | 2177.8496 | 275.09167 | 559.3206 | NA | 111.24314 | 501.2060 | 105982.9 | 10663.714 | 15022.21 | NA | 613.3968 | 1224.3837 | 946.0795 | 309.7599 | 270.67770 | 1808.1924 | 21555.3555 | 200.7485 | 342.1992 | 1227.435 | 26587.62 | 13723.719 | 22957.35 | 551.6828 | 1176.7791 | 319.0364 | NA | 118.5104 | 3499.113 | 26124.20 | 91.82145 | 319.1320 | 1003.3372 | 1342.4712 | 143.12419 | 1352.7024 | 430.13318 | 144.6799 | 171.13177 | 221.9161 | 1889.0665 | 835.6825 | NA |
| 100pmol | 4 | 1620.490 | 3828.19971 | 2062.8384 | 385.05573 | 558.0967 | 422.0465 | 84.27336 | 334.6389 | 104442.6 | 10843.115 | 15160.49 | NA | 886.5406 | 1148.7343 | 1091.7800 | NA | 229.40149 | 901.5703 | 22937.2500 | 240.7981 | 418.1846 | 1190.952 | 26168.72 | 13944.603 | 22311.30 | 438.5425 | 1162.6656 | 351.5390 | NA | 137.8860 | 3481.821 | 25910.39 | 88.26187 | 217.7478 | 489.8084 | 1721.8601 | 99.95578 | 990.6649 | 393.55930 | 134.5238 | 145.17339 | 216.3736 | 1610.2407 | 950.3087 | 913.3416 |
| 200pmol | 1 | 1512.770 | 4232.05078 | 2004.8613 | 338.27777 | 156.3478 | 364.5416 | 146.80331 | NA | 109245.3 | 19524.863 | 21577.97 | 2212.190 | 491.7787 | 1246.4460 | 1080.4132 | 270.1487 | 252.09808 | 1454.3271 | 21113.4512 | 223.8396 | 313.7860 | 1176.982 | 48693.35 | 24344.188 | 41234.67 | 364.7307 | 1203.0853 | 385.5154 | 65.40555 | 151.0895 | 3553.484 | 26261.47 | 81.22160 | 185.4865 | 939.8899 | 2149.7632 | 131.13179 | 381.0588 | 429.62201 | 239.4998 | 145.04378 | 424.7914 | 2337.8496 | NA | 837.8737 |
| 200pmol | 2 | 1480.490 | 3496.84155 | 2177.9534 | NA | 550.4083 | NA | 135.78349 | 295.8571 | 113357.5 | 20072.297 | 22968.96 | NA | 669.7894 | 1068.2001 | NA | 285.4891 | 259.50000 | 1049.7526 | 25760.0527 | 190.3054 | 452.8294 | 1220.266 | 49866.29 | 24742.227 | 42899.43 | 633.5656 | 1234.5601 | 414.1271 | NA | 135.8605 | 3686.869 | 27638.89 | 69.56509 | 250.4035 | 1020.4291 | 725.6615 | 116.20615 | 877.0164 | 438.22589 | 133.4297 | 160.92671 | 155.0986 | NA | 1053.8444 | 1000.5491 |
| 200pmol | 3 | 1555.834 | 356.43225 | 2280.6846 | 379.62103 | 564.2863 | 496.0772 | 103.30424 | 473.9141 | 114321.8 | 20787.127 | 20720.13 | 1451.198 | 586.7260 | 1378.0652 | 1194.8448 | 291.6754 | 184.18954 | 1123.7469 | NA | 174.5702 | 432.1681 | 1216.306 | 50704.73 | 24803.633 | 42904.95 | 446.4135 | 1082.7312 | 357.6343 | NA | 129.0676 | 3530.710 | 27101.22 | 62.08423 | 136.7023 | 1171.5715 | 1675.6870 | 109.60301 | 938.3956 | 568.89239 | 315.7039 | 146.75146 | 198.4779 | 1397.9890 | 837.2197 | 694.5791 |
| 200pmol | 4 | 1529.628 | 350.70822 | 2223.3093 | 410.82349 | 292.9041 | 522.1325 | 95.18819 | 318.4948 | 116439.8 | 19924.240 | 22153.40 | NA | 539.0703 | 923.3237 | 1115.3848 | 322.9086 | 97.65465 | 957.0436 | NA | 164.7767 | NA | 1183.197 | 53744.70 | 26381.047 | 43279.84 | 527.1628 | 1121.3438 | 342.5055 | NA | 121.3068 | 3751.769 | 27545.24 | 70.39470 | 199.2453 | 996.0696 | 1696.6189 | 125.31519 | 611.6407 | 506.49115 | 204.4332 | 161.96100 | 376.5362 | 895.9138 | NA | NA |
| 50pmol | 1 | 1480.210 | 561.38837 | 189.9275 | 264.24271 | 308.9420 | NA | 599.90497 | 192.3859 | 117803.5 | 6758.298 | 12183.81 | NA | 594.8999 | 899.5010 | 1163.1122 | 291.4431 | 176.21545 | 620.2048 | 14107.1250 | 152.5492 | 292.2440 | 1186.543 | 16408.28 | 7169.955 | 14728.67 | 2984.7190 | 1029.7336 | 288.4770 | 891.24725 | 129.7482 | 3547.950 | 25668.78 | 846.95880 | 146.3040 | NA | 461.3821 | 86.84789 | 373.6308 | 49.93938 | 236.2902 | 20.92994 | 142.3466 | NA | NA | NA |
| 50pmol | 2 | 1486.144 | NA | 1462.2559 | 325.74991 | 351.2331 | NA | 254.75084 | 308.6775 | 110086.7 | 6721.135 | 12521.78 | NA | 582.8912 | 531.7106 | 1119.5256 | 287.1180 | 103.58258 | 849.2368 | 24912.3613 | 140.6493 | 362.3117 | 1260.574 | 16444.63 | 7797.536 | 14736.71 | 857.5026 | NA | 361.4482 | 179.10303 | 166.8891 | 3530.004 | 26351.25 | 207.83086 | 165.6463 | 265.2173 | 1184.9562 | 93.91448 | 768.2026 | 489.40918 | 146.9422 | 88.41573 | 101.6087 | NA | NA | NA |
| 50pmol | 3 | 1468.554 | 42.51457 | 1364.9075 | 83.99377 | 296.5147 | 396.0038 | 257.78970 | 279.2477 | 105640.2 | 6172.877 | 11926.22 | 1373.660 | 569.8922 | NA | 1067.0791 | 294.0919 | 88.48861 | 738.7719 | 666.5015 | NA | NA | 1175.953 | 16618.11 | 7432.793 | 14160.20 | 916.4893 | 992.5451 | 319.6350 | 128.63672 | 120.6974 | 3458.023 | 26017.54 | 203.64948 | 132.5755 | 291.4759 | 932.9668 | 93.50905 | 547.0935 | 263.86734 | 313.0341 | 111.88376 | 85.4563 | NA | NA | NA |
| 50pmol | 4 | 1497.531 | 927.07886 | 1435.5588 | 275.60831 | 242.4643 | 425.7305 | 197.71338 | 382.4084 | 110446.0 | 6028.398 | 12021.50 | NA | NA | 593.1353 | 1302.1250 | 339.3387 | 30.13688 | 873.1840 | 15711.3106 | 142.4270 | 291.5121 | 1150.711 | 16282.51 | 7543.633 | 14758.73 | 886.7808 | 1138.6193 | NA | 152.56187 | NA | 3575.316 | 25969.99 | 190.47060 | 220.1901 | 676.8246 | 996.8993 | 31.57284 | 523.4712 | 450.08408 | 164.1874 | 143.96025 | 135.2896 | NA | NA | NA |
Details
The function preprocessing() takes a .csv
file of summarized protein abundances, exported from
Spectronaut. The most important columns that need to be
included in this file are: R.Condition,
R.Replicate, PG.ProteinAccessions,
PG.ProteinNames,
PG.NrOfStrippedSequencesIdentified, and
PG.Quantity. This function will reformat the data and
provide functionality for some initial filtering (based on the number of
unique peptides). The steps below describe the functions that happen in
the Preprocessing code.
1. Loads the raw data
If the raw data is in a .csv file Toy_Spectronaut_Data.csv, specify the
fileNameto read the raw data file into R.If the raw data is stored as an .RData file Toy_Spectronaut_Data.RData, first load the data file directly, then specify the
dataSetin the function.
2. Filters out identified proteins that exhibit “NaN” quantitative values
NaN, which stands for ‘Not a Number,’ can be found in the PG.Quantity column for proteins that were identified by MS and MS/MS evidence in the raw data, but all peptides from that protein lack an associated integrated peak area or intensity. This usually occurs in low abundance peptides that exhibit intensities close to the limit of detection resulting in poor signal-to-noise (S/N) and/or when there is interference from other co-eluting peptide ions with very similar or identical m/z values that lead to difficulty in parsing out individual intensity profiles.
3. Applies a unique peptides per protein filter
General practice in the proteomics field is to filter out proteins which were identified on the basis of a single peptide. Because approximately 1% of all identified peptides are false positive matches, it’s more likely that 1 peptide was incorrectly identified and that protein ID is incorrect than that, for example, 5 peptides from the same protein were all incorrectly identified and that protein ID is incorrect. We recommend focusing on proteins with 2 or more peptide identifications, as these will be higher confidence. If you have a protein of interest with only 1 peptide identified, contact PMF faculty and we can help you evaluate the evidence from the raw data to determine believability.
4. Adds accession numbers to identified proteins without informative names
Spectronaut reports contain 4 different columns of identifying information:
-
PG.Genes, which is the gene name (e.g. CDK1). -
PG.ProteinAccessions, which is the UniProt identifier number for a unique entry in the online database (e.g. P06493). -
PG.ProteinDescriptions, which is the protein name as provided on UniProt (e.g. cyclin-dependent kinase 1). -
PG.ProteinNames, which is a concatenation of an identifier and the species (e.g. CDK1_HUMAN).
Every entry in UniProt will have an accession number, but may not
have all of the other identifiers, due to incomplete annotation. Because
Uniprot includes entries for fragments of proteins and some proteins
entries are redundant, a peptide can match to multiple entries for the
same protein, which generates multiple possible identifiers in
Spectronaut. Further, the ProteinNames
entry in Spectronaut can switch formats: the preference
is accession number and species, but can also be gene name and species
instead.
This option tells msDiaLogue to substitute the accession number for an identifier if it tries to pull an identifier from a column with no information.
5. Saves a document to your working directory with all filtered out data, if desired
If saveRm = TRUE, the data removed in step 2
(preprocess_filterNaN.csv) and step 3
(preprocess_filterUnique.csv) will be saved in the current
working directory.
As part of the preprocessing(), a histogram of
-transformed
protein abundances is provided. This is a helpful way to confirm that
the data have been read in correctly, and there are no issues with the
numerical values of the protein abundances. Ideally, this histogram will
appear fairly symmetrical (bell-shaped) without too much skew towards
smaller or larger values.